Tutorials
Note
Click on the repository link to download the zip file, called tutorialData.zip, of the data you’ll need to run through the tutorials.
Make a working directory and do the tutorials in that directory.
Note
In the gif above, the working directory is located at Documents/demos/. The tutorial data files were placed in the directory called “demos.” Whatever you call it, when working in the terminal, you’ll always need to be in that directory to complete the tutorial.
The gif also showed a few commands that you may find helpful:
pwdto find the directory you’re currently in
cdto change directories
lsto list files in the folder
This tutorial builds up! Between each example and the previous examples, the new lines of code are highlighted in yellow.
Python
Python is an interpreted object oriented programming language. There is a large range of modules that are imported into python to provide extra functionality or features. pyWitness uses numpy (numerical arrays), scipy (fitting and functions), pandas (data frames), matplotlib (plotting), openpyxl (reading/writing excel), xlrd (reading/writing excel), and numba (compiler to speed up code).
Python is best started from a terminal/command prompt
ipython3 --pylab
This then lands you in a python console window
Python 3.7.9 (default, Sep 6 2020, 16:32:30)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.14.0 -- An enhanced Interactive Python. Type '?' for help.
In [1]:
Commands can now be typed in to execute python and pyWitness commands. Here are some helpful tips to speed up inputing commands
Cut and paste commands (to reduce typos)
Use the command history (up and down cursor arrows) to find commands that were used previously
Use command history with search (so try
import pyWand then up arrow. This will search the command history with that command fragment and probably match with a previousimport pyWitnessA command can be completed by using
tab. Try typing inimport pyWand then pressingtabTo get help on a command, type the function and then
?for example,dp.plotROC?
Loading raw experimental data
Remember, you may need to activate pyWitness when you start a terminal by using this code
conda activate pyWitness
Start up ipython3 with
ipython3 --pylab
and pyWitness with
import pyWitness
A single Python class pyWitness.DataRaw is used to load raw data in
either csv or excel format. The format of test1.csv is the same as that described in the introduction.
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
pyw <- import("pyWitness")
Checking and exploring loaded data
It is useful to understand what columns and data values are stored in the raw data.
1 2 3 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.checkData() |
1 2 3 | pyw <- pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$checkData() |
DataRaw.checkData>
DataRaw.checkData> columns : ['Unnamed: 0' 'participantId' 'lineupSize' 'targetLineup' 'responseType' 'confidence' 'responseTime']
DataRaw.checkData> lineupSize : [6]
DataRaw.checkData> targetLineup : ['targetAbsent' 'targetPresent']
DataRaw.checkData> responseType : ['fillerId' 'rejectId' 'suspectId']
DataRaw.checkData> confidence : [ 0 10 20 30 40 50 60 70 80 90 100]
DataRaw.checkData> number trials : 890
If the unique values for a non-mandatory column are required then this can be displayed using
1 2 3 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.columnValues("responseTime") |
1 2 3 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$columnValues("responseTime") |
DataRaw.columnValues> : responseTime [ 1159 1296 1326 ... 161703 502420 651073]
It is possible also to load Excel files
import pyWitness
dr = pyWitness.DataRaw("test1.xlsx","test1")
pyw <- import("pyWitness")
dr <- pyw$DataRaw("test1.xlsx","test1")
The second argument is the sheet name within the workbook (in the example above, it’s “test1”).
Processing raw experimental data
To process the raw data the function pyWitness.DataRaw.process needs to be called on a raw data object. This calculates the cumulative rates from the raw data.
1 2 3 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dp = dr.process() |
1 2 3 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() |
Once pyWitness.DataRaw.process is called two DataFrames are
created. One contains a pivot table and the other contains rates.
1 2 3 4 5 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dp = dr.process() dp.printPivot() dp.printRates() |
1 2 3 4 5 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() dp$printPivot() dp$printRates() |
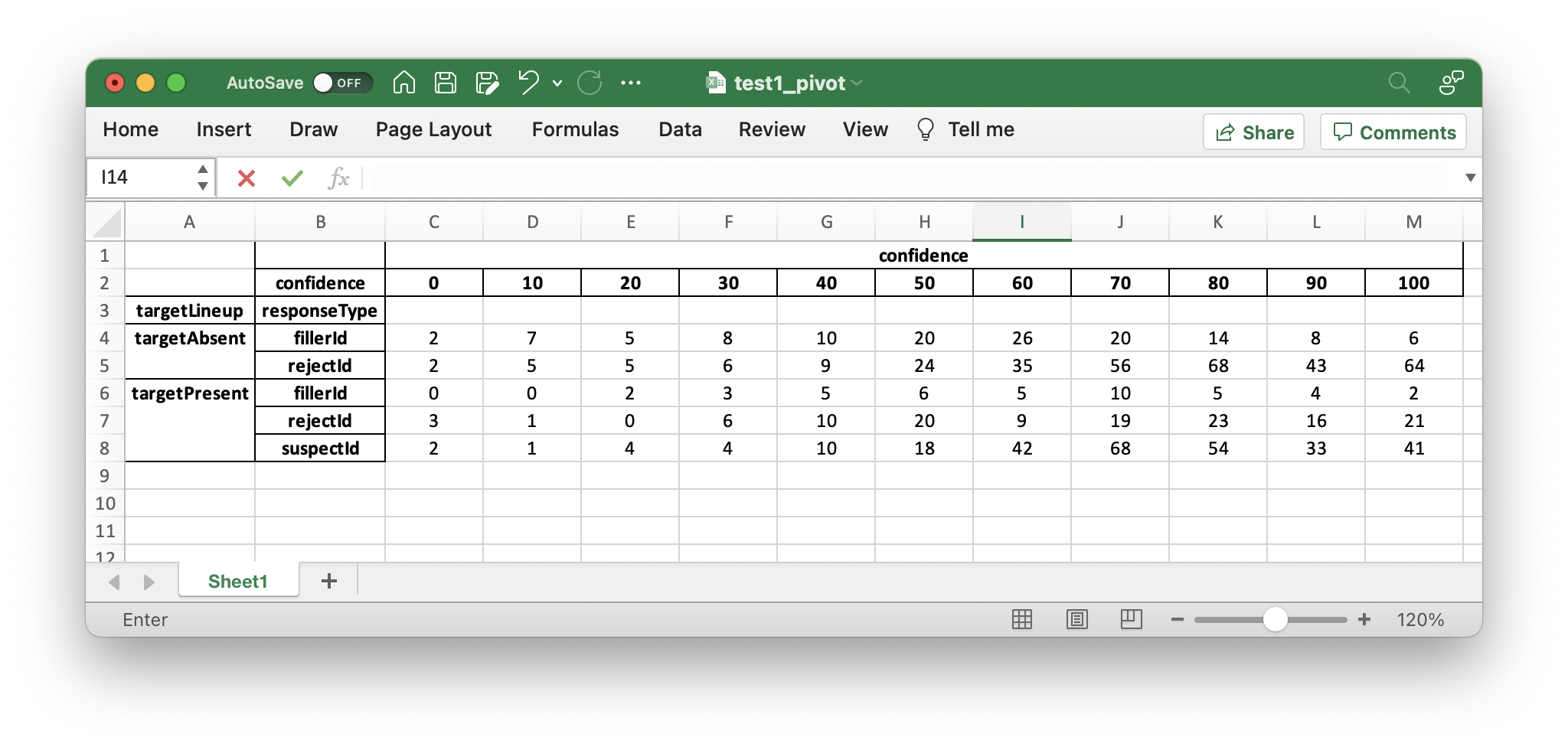
You should see the following output of the dp.printPivot().
confidence
confidence 0 10 20 30 40 50 60 70 80 90 100
targetLineup responseType
targetAbsent fillerId 2.0 7.0 5.0 8.0 10.0 20.0 26.0 20.0 14.0 8.0 6.0
rejectId 2.0 5.0 5.0 6.0 9.0 24.0 35.0 56.0 68.0 43.0 64.0
targetPresent fillerId 0.0 0.0 2.0 3.0 5.0 6.0 5.0 10.0 5.0 4.0 2.0
rejectId 3.0 1.0 0.0 6.0 10.0 20.0 9.0 19.0 23.0 16.0 21.0
suspectId 2.0 1.0 4.0 4.0 10.0 18.0 42.0 68.0 54.0 33.0 41.0
total number of participants 890.0
In the output above are frequencies by confidence levels for each response type. To familiarize you with the output, in the table above, 8 filler identifications were given with 30% confidence on the target-absent lineups, 64 reject identifications (i.e., “The perp is not in the lineup”) given with 100% confidence on the target-absent lineups, and 41 guilty suspect identifications (from target-present lineups) given with 100% confidence.
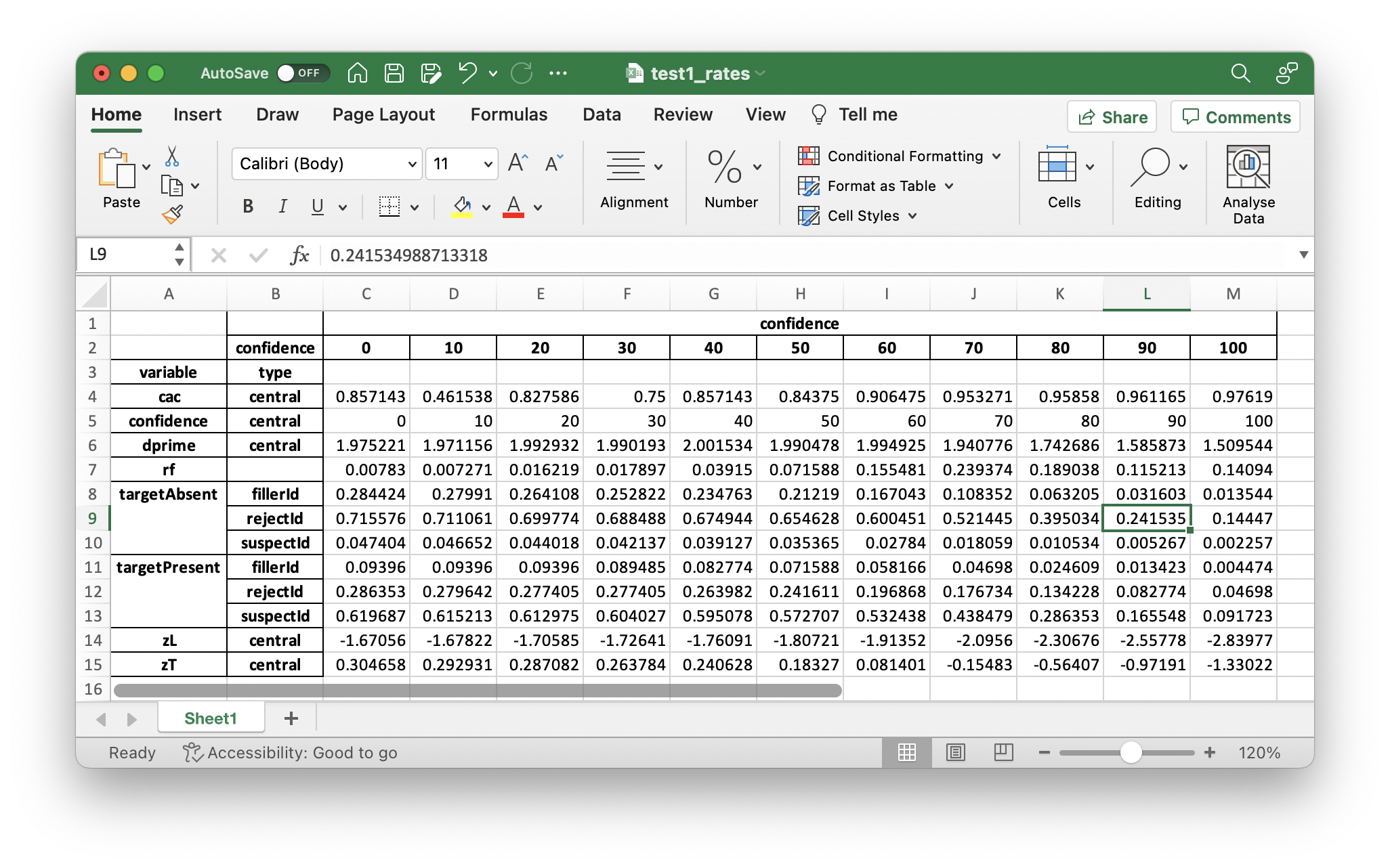
You should also see the following output for dp.printRates()
confidence
confidence 0 10 20 30 40 50 60 70 80 90 100
variable type
cac central 0.857143 0.461538 0.827586 0.750000 0.857143 0.843750 0.906475 0.953271 0.958580 0.961165 0.976190
confidence central 0 10 20 30 40 50 60 70 80 90 100
dprime central 1.975221 1.971156 1.992932 1.990193 2.001534 1.990478 1.994925 1.940776 1.742686 1.585873 1.509544
rf 0.007830 0.007271 0.016219 0.017897 0.039150 0.071588 0.155481 0.239374 0.189038 0.115213 0.140940
targetAbsent fillerId 0.284424 0.279910 0.264108 0.252822 0.234763 0.212190 0.167043 0.108352 0.063205 0.031603 0.013544
rejectId 0.715576 0.711061 0.699774 0.688488 0.674944 0.654628 0.600451 0.521445 0.395034 0.241535 0.144470
suspectId 0.047404 0.046652 0.044018 0.042137 0.039127 0.035365 0.027840 0.018059 0.010534 0.005267 0.002257
targetPresent fillerId 0.093960 0.093960 0.093960 0.089485 0.082774 0.071588 0.058166 0.046980 0.024609 0.013423 0.004474
rejectId 0.286353 0.279642 0.277405 0.277405 0.263982 0.241611 0.196868 0.176734 0.134228 0.082774 0.046980
suspectId 0.619687 0.615213 0.612975 0.604027 0.595078 0.572707 0.532438 0.438479 0.286353 0.165548 0.091723
zL central -1.670562 -1.678225 -1.705849 -1.726409 -1.760906 -1.807208 -1.913524 -2.095603 -2.306755 -2.557781 -2.839765
zT central 0.304658 0.292931 0.287082 0.263784 0.240628 0.183270 0.081401 -0.154827 -0.564069 -0.971908 -1.330222
In the table above, the overall false ID rate is 0.047, the overall correct ID rate is 0.620, and the overall correct rejection rate is 0.716.
Note
In the example there is no suspectId for targetAbsent lineups. Here the targetAbsent.suspectId is estimated as targetAbsent.fillerId/lineupSize
To see overall descriptive statistics, use
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
dp = dr.process()
dp.printDescriptiveStats()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dp <- dr$process()
dp$printDescriptiveStats()
and you’ll see this output:
Number of lineups 890.0
Number of target-absent lineups 443.0
Number of target-present lineups 447.0
Correct ID rate 0.6196868008948546
False ID rate 0.0474040632054176
dPrime 1.9752208100241062
pAUC 0.02066155955774986
Plotting ROC curves
1 2 3 4 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dp = dr.process() dp.plotROC() |
1 2 3 4 5 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() dp$plotROC() mpl$pyplot$show() |
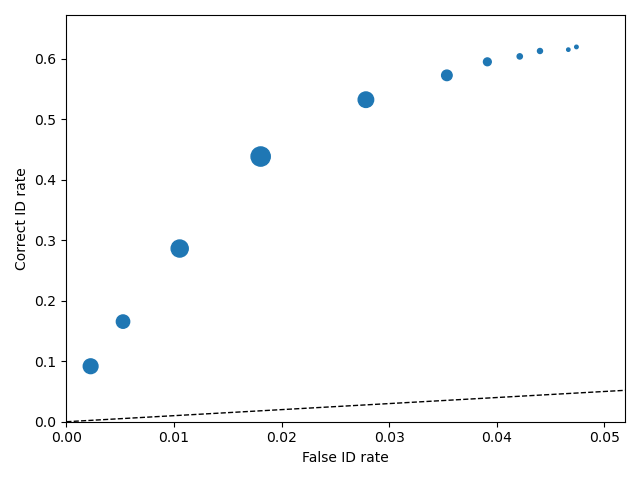
Note
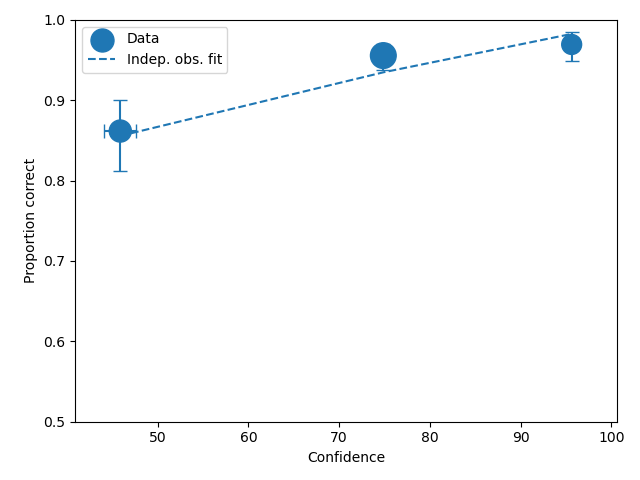
The symbol size is the relative frequency and can be changed by setting dp.plotROC(relativeFrequencyScale = 400)
Note
The transparency of the plot can be changed by setting alpha in the plot command, so dp.plotROC(alpha = 0.5)
The black dashed line in the plot represents chance performance.
Plotting CAC curves
1 2 3 4 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dp = dr.process() dp.plotCAC() |
1 2 3 4 5 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() dp$plotCAC() mpl$pyplot$show() |
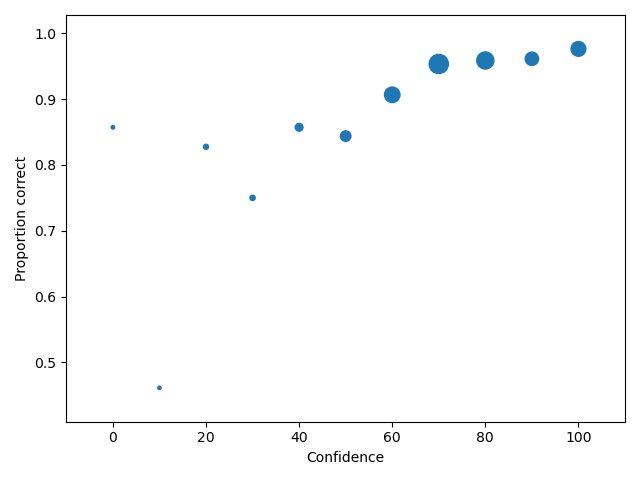
Note
The transparency of the plot can be changed by setting alpha in the plot command, so dp.plotCAC(alpha = 0.5)
Warning
To plot CAC curves with different target-present base rates, the base rate needs to be given to the process function of DataRaw, for example, dp = dr.process(baseRate=0.2)
Collapsing the categorical data
The dataset used in this tutorial has 11 confidence levels (0, 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100). Often confidence levels need to be binned or collapsed. This is best performed on the raw data before calling
process(). This is done with the collapseCategoricalData method of DataRaw, and shown in example below, where the new bins are (0-60 map to 30, 70-80 to 75 and 90-100 to 95).
1 2 3 4 5 6 7 8 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseCategoricalData(column='confidence', map={0: 30, 10: 30, 20: 30, 30: 30, 40: 30, 50: 30, 60: 30, 70: 75, 80: 75, 90: 95, 100: 95}) dp = dr.process() dp.plotCAC() |
1 2 3 4 5 6 7 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseCategoricalData(column='confidence',map=list("0"=30, "10"=30, "20"=30, "30"=30, "40"=30,"50"=30, "60"=30,"70"=75, "80"=75,"90"=95, "100"=95)) dp <- dr$process() dp$plotCAC() mpl$pyplot$show() |
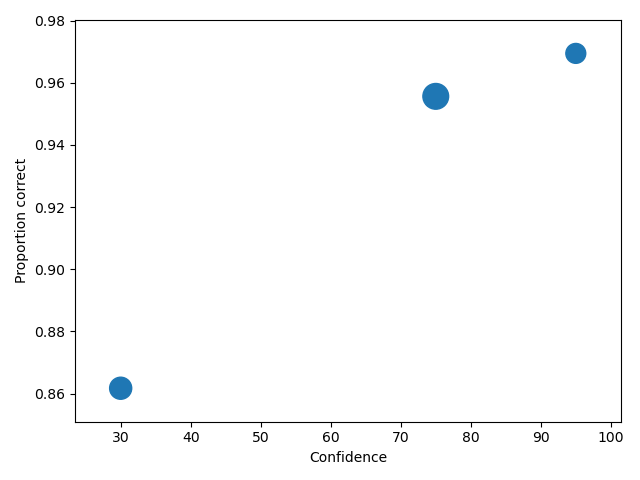
To rescale the axes, you can use
import matplotlib as _plt
xlim(0,100)
ylim(0.50,1.0)
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseCategoricalData(column='confidence',map=list("0"=30, "10"=30, "20"=30, "30"=30, "40"=30,"50"=30, "60"=30,"70"=75, "80"=75,"90"=95, "100"=95))
dp <- dr$process()
dp$plotCAC()
invisible(mpl$pyplot$ylim(0.50,1.0))
mpl$pyplot$show()
and you get
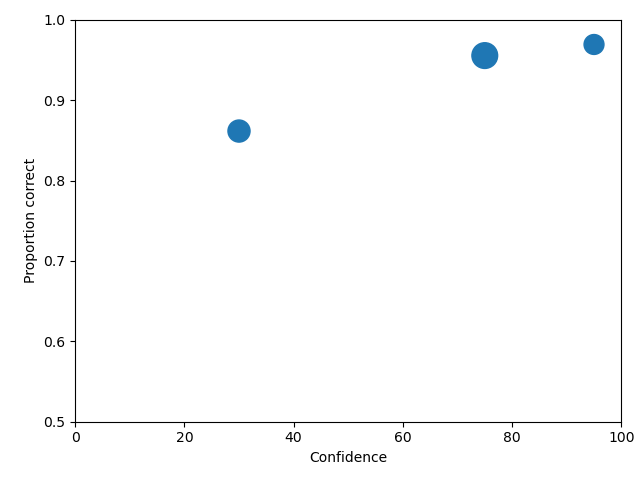
Note
If you err, the collapseCategoricalData the data might be inconsistent. To start with the original data so call collapseCategoricalData with reload=True
Collapsing (binning) continuous data
Some data are not categorical variables, but continuous variables.
1 2 3 4 5 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() dp.plotROC() |
1 2 3 4 5 6 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence", bins = c(-1,60,80,100),labels= c(1,2,3)) dp <- dr$process() dp$plotROC() mpl$pyplot$show() |
Note
labels=None can be used and the bins will be automatically labelled
Note
The bin edges are exclusive of the low edge and inclusive of the high edge
Warning
Confidence needs to be a numerical value because ROC analysis requires a value that can be ordered.
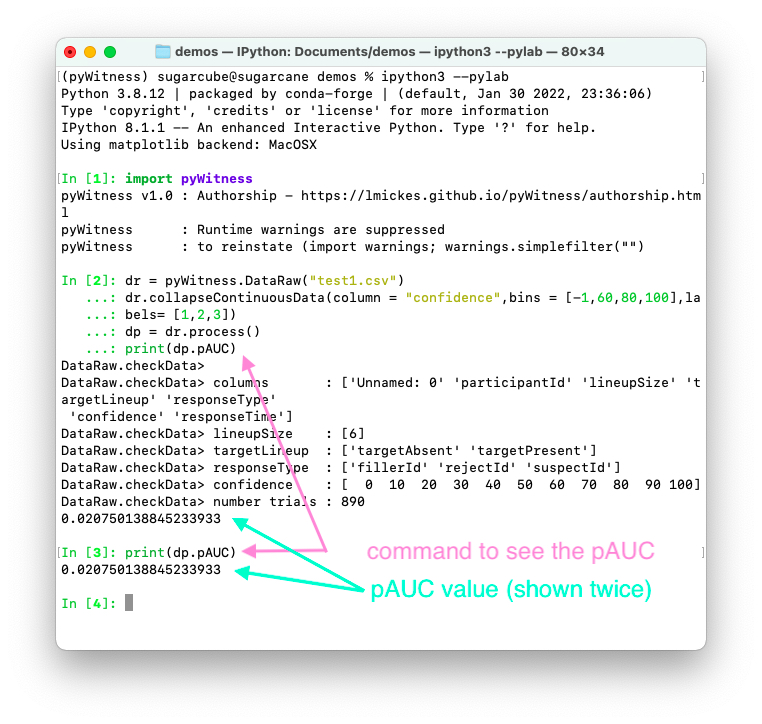
Calculating pAUC and performing statistical tests
pAUC is calculated when dr.process() is called. Simpson’s rule integrates the area
under the ROC curve up to a maximum value. If the maximum value is between two data points, linear interpolation is used to calculate the most liberal point (i.e., the lowest level of confidence).
1 2 3 4 5 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() print(dp.pAUC) |
1 2 3 4 5 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels= c(1,2,3)) dp <- dr$process() print(dp$pAUC) |
Plotting RAC curves
To perform analyses with a different variable than confidence, for example, response time, use the following code. The important change is highlighted.
1 2 3 4 5 | import pyWitness drRAC = pyWitness.DataRaw("test1.csv") drRAC.collapseContinuousData(column="responseTime",bins=[0, 5000, 10000, 15000, 20000, 99999],labels=[1, 2, 3, 4, 5]) dpRAC = drRAC.process(reverseConfidence=True,dependentVariable="responseTime") dpRAC.plotCAC() |
1 2 3 4 5 6 7 8 9 10 | pyw <- import("pyWitness") drRAC <- pyw$DataRaw("./test1.csv") drRAC$collapseContinuousData(column="responseTime", bins=c(0, 5000, 10000, 15000, 20000, 99999),labels=c(1, 2, 3, 4, 5)) dpRAC <- drRAC$process(reverseConfidence=TRUE,dependentVariable="responseTime") dpRAC$plotCAC() invisible(mpl$pyplot$xlabel("Response Time")) invisible(mpl$pyplot$ylim(.50,1.0)) invisible(mpl$pyplot$savefig("test1RAC.png")) invisible(mpl$pyplot$savefig("test1RAC.pdf")) |
The plot will look like this:

Fitting signal detection-based models to data
There are many models available in pyWitness. We’ll start with the independent observation model. To load and process the data is the same as before (lines 1-4), the fitting part is new and the code is highlighted (lines 5-7).
1 2 3 4 5 6 7 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() mf = pyWitness.ModelFitIndependentObservation(dp) mf.setEqualVariance() mf.fit() |
1 2 3 4 5 6 7 | pyw <- import("pyWitness",convert=TRUE) dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels= c(1,2,3)) dp <- dr$process() mf <- pyw$ModelFitIndependentObservation(dp) mf$setEqualVariance() mf$fit() |
Line 5 constructs a fit object, line 6 sets the model parameters to equal variance and line 7 starts the minimiser. The output from the fit (execution of line 7) is something like the following
fit iterations 223
fit status Optimization terminated successfully.
fit time 9.376720442
fit chi2 10.300411274463407
fit ndf 4
fit chi2/ndf 2.5751028186158518
fit p-value 0.035660197825222784
To clearly see how the fitting works, the following code is the same as above but
with mf.printParameters() on lines 6, 9, and 12.
1 2 3 4 5 6 7 8 9 10 11 12 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() mf = pyWitness.ModelFitIndependentObservation(dp) mf.printParameters() mf.setEqualVariance() mf.printParameters() mf.fit() mf.printParameters() |
1 2 3 4 5 6 7 8 9 10 11 12 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels= c(1,2,3)) dp <- dr$process() mf <- pyw$ModelFitIndependentObservation(dp) mf$printParameters() mf$setEqualVariance() mf$printParameters() mf$fit() mf$printParameters() |
After creating the mf object (line 9) the parameters are at their default values and free
lureMean 0.0 (free)
lureSigma 1.0 (free)
targetMean 1.0 (free)
targetSigma 1.0 (free)
lureBetweenSigma 0.0 (free)
targetBetweenSigma 0.0 (free)
c1 1.0 (free)
c2 1.5 (free)
c3 2.0 (free)
Typically you would want to control the fit parameters. setEqualVariance sets some default model which is
an appropriate start; line 12 yields
lureMean 0.0 (fixed)
lureSigma 1.0 (fixed targetSigma)
targetMean 1.0 (free)
targetSigma 1.0 (fixed)
lureBetweenSigma 0.3 (fixed targetBetweenSigma)
targetBetweenSigma 0.3 (free)
c1 1.0 (free)
c2 1.5 (free)
c3 2.0 (free)
Comparing these two fit parameters settings
lureSigmais forced to be equal totargetSigma
targetSigmais fixed to its current value
lureBetweenSigmais fixed totargetBetweenSigma
targetBetweenSigmais fixed to its current value
After running the fit the parameters are updated so the output of line 12 in the code example gives
ModelFit.printParameters> lureMean 0.000 (fixed)
ModelFit.printParameters> lureSigma 1.000 (fixed targetSigma)
ModelFit.printParameters> targetMean 1.798 (free)
ModelFit.printParameters> targetSigma 1.000 (fixed)
ModelFit.printParameters> lureBetweenSigma 0.605 (fixed targetBetweenSigma)
ModelFit.printParameters> targetBetweenSigma 0.605 (free)
ModelFit.printParameters> c1 1.402 (free)
ModelFit.printParameters> c2 1.935 (free)
ModelFit.printParameters> c3 2.677 (free)
There many ways to control the model
Command |
Notes |
|---|---|
|
Sets the lure mean parameter to -0.1 |
|
Fixed the parameter so it cannot change during a fit |
|
Unfixes the parameter so it will be free in a fit |
|
Locks |
|
Release the linking of lureBetweenSigma and targetBetweenSigma |
There are multiple fits available and they all have the same interface but differ in the construction line
1 2 3 4 5 6 7 8 | dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column="confidence") dp = dr.process() mf_io = pyWitness.ModelFitIndependentObservation(dp) mf_br = pyWitness.ModelFitBestRest(dp) mf_en = pyWitness.ModelFitEnsemble(dp) mf_in = pyWitness.ModelFitIntegration(dp) |
1 2 3 4 5 6 7 8 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() mf_io <- pyw$ModelFitIndependentObservation(dp) mf_br <- pyw$ModelFitBestRest(dp) mf_en <- pyw$ModelFitEnsemble(dp) mf_in <- pyw$ModelFitIntegration(dp) |
Setting initial fit parameters
With data samples with large number of confidence bins the fits can take a large number of iterations to converge (long run times). Sensible fit parameters can be be estimated from the data.
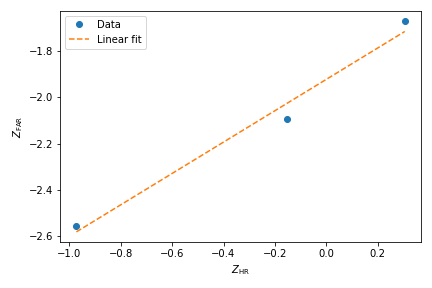
To estimate the target mean \(\mu_t\) and sigma \(\sigma_t\) the following relation is used
Rearranging gives
There is a linear relationship between target and lure \(Z\) values. This can be plotted and a linear fit used to estimate the gradient and intercept.
1 2 3 4 5 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() dp.plotHitVsFalseAlarmRate() |
1 2 3 4 5 6 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels = c(1,2,3)) dp <- dr$process() dp$plotHitVsFalseAlarmRate() invisible(mpl$pyplot$savefig("HvFA.png")) |
1 2 3 4 5 6 7 8 9 10 11 12 13 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= [1,2,3]) dp = dr.process() mf = pyWitness.ModelFitIndependentObservation(dp) mf.printParameters() mf.setEqualVariance() mf.setParameterEstimates() mf.printParameters() mf.fit() mf.printParameters() |
1 2 3 4 5 6 7 8 9 10 11 12 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels = c(1,2,3)) dp <- dr$process() mf <- pyw$ModelFitIndependentObservation(dp) mf$setEqualVariance() mf$setParameterEstimates() mf$printParameters() mf$fit() mf$printParameters() |
Plotting fit and models
It is important to understand the performance of a given particular fit. The following plot compares the experimental data to the model fit.
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels= None)
dp = dr.process()
dp.calculateConfidenceBootstrap(nBootstraps=200)
mf = pyWitness.ModelFitIndependentObservation(dp)
mf.setEqualVariance()
mf.fit()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels = c(1,2,3))
dp <- dr$process()
dp$calculateConfidenceBootstrap(nBootstraps=as.integer(200))
mf <- pyw$ModelFitIndependentObservation(dp)
mf$setEqualVariance()
mf$fit()
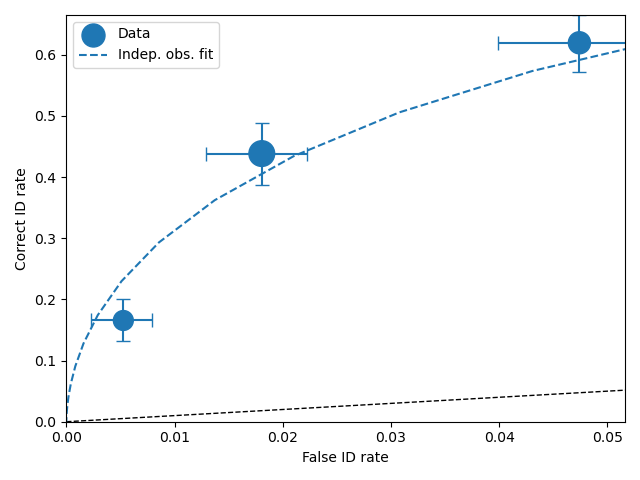
To compare an ROC plot between data and fit
dp.plotROC(label="Data")
mf.plotROC(label="Indep. obs. fit")
import matplotlib.pyplot as _plt
_plt.legend()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels = c(1,2,3))
dp <- dr$process()
dp$calculateConfidenceBootstrap(nBootstraps=as.integer(200))
mf <- pyw$ModelFitIndependentObservation(dp)
mf$setEqualVariance()
mf$fit()
dp$plotROC(label="Data")
mf$plotROC(label="Indep. obs. fit")
mpl$pyplot$legend()
mpl$pyplot$show()
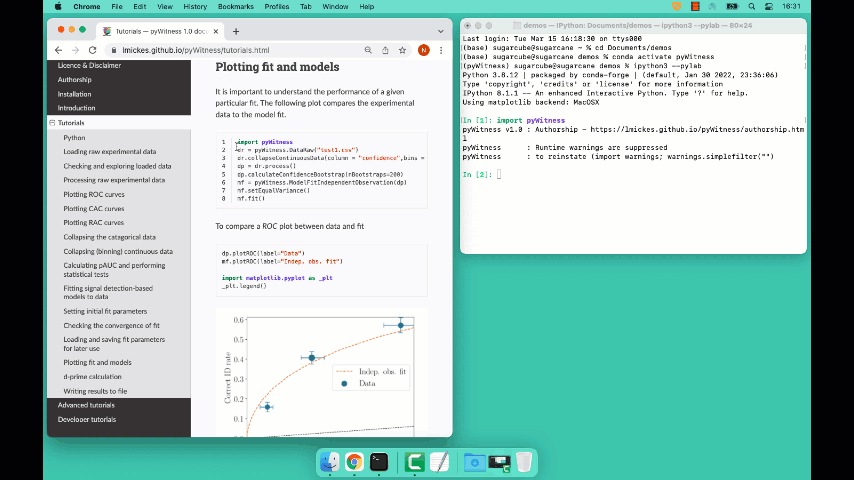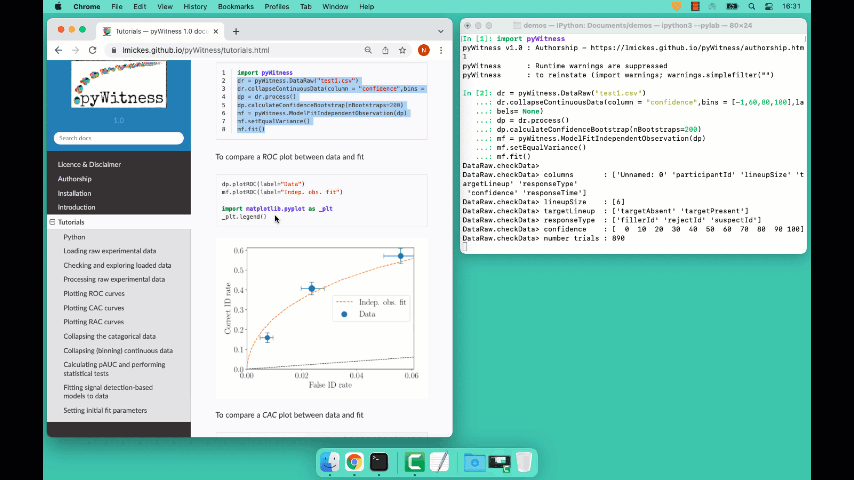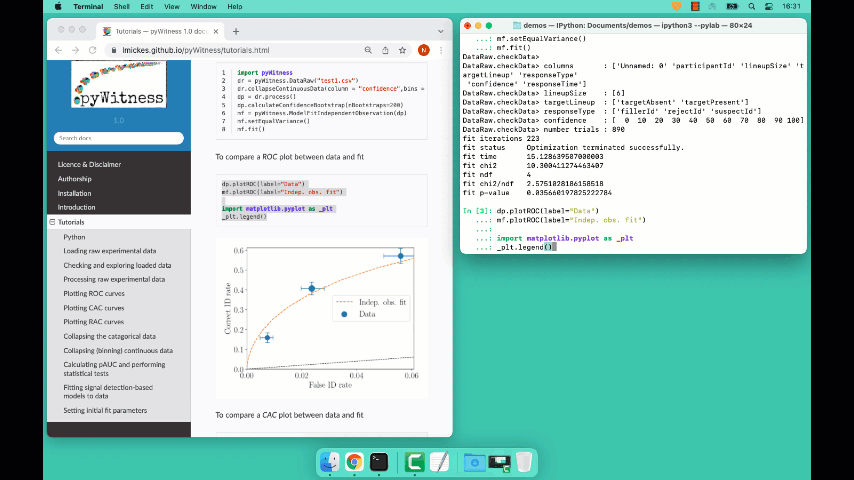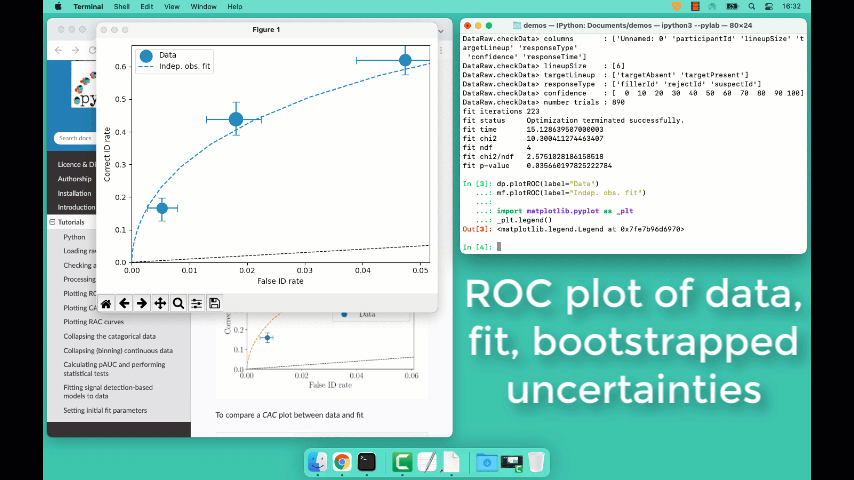
To compare a CAC plot between data and fit
dp.plotCAC(label="Data")
mf.plotCAC(label="Indep. obs. fit")
import matplotlib.pyplot as _plt
_plt.legend()
dp$plotCAC(label="Data")
mf$plotCAC(label="Indep. obs. fit")
mpl$pyplot$legend()
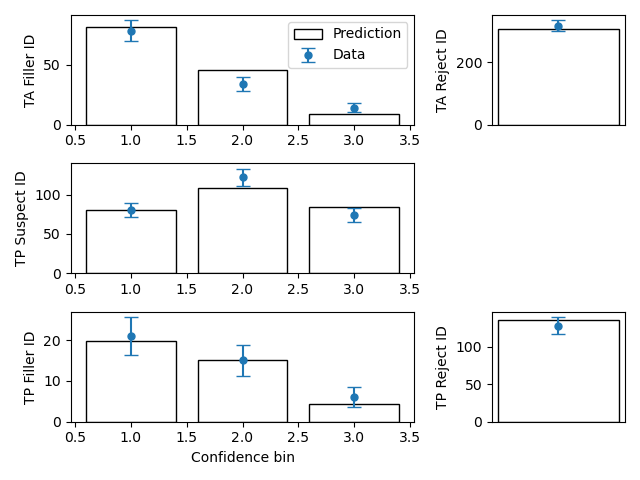
To compare frequencies in each bin between data and fit
mf.plotFit()
mf$plotFit()
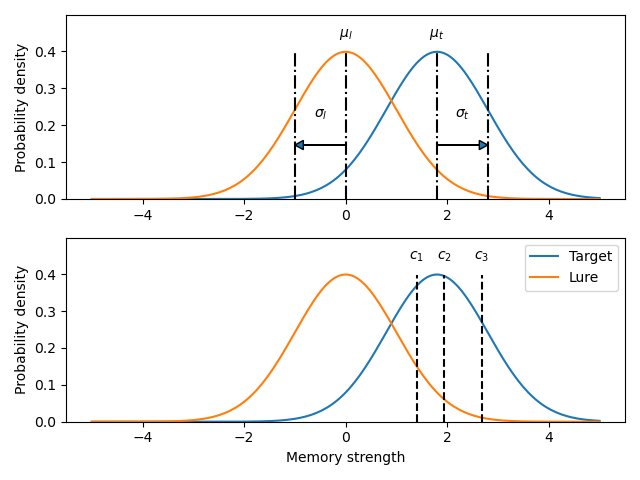
Once a fit has been performed, the model can be displayed as a function of memory strength and includes the lure and target distributions with means and standard deviations (top panel of plot below) and the associated criteria, c1 (low confidence), c2 (medium confidence), and c3 (high confidence) (bottom panel of plot below). This simple command belonging to a ModelFit object can be used to make the plot below.
mf.plotModel()
mf$plotModel()
d-prime calculation
The d-prime can be calculated by computing
where \(R_{T,i}\) is the cumulative rate for targets (\(T\)) with confidence \(i\), \(R_{L,i}\) is the cumulative
rate for lures (\(L\)) with confidence \(i\) and \(Z\) is the inverse normal CDF. This can be evaluated for every
confidence bin, but there are conventions for lineups and showups. For all confidence levels \(d^{\prime}\) is stored in the rates
dataframe, so dp.printRates() gives
1 2 3 4 5 6 7 8 9 10 11 12 13 | confidence confidence 3 2 1 targetLineup responseType cac central 0.956357 0.940618 0.839228 confidence central 95.588235 74.859335 44.778068 dprime central 1.433207 1.748223 1.767339 rf 0.264691 0.422903 0.312406 targetAbsent fillerId 0.044660 0.141748 0.335922 rejectId 0.217476 0.473786 0.664078 suspectId 0.007443 0.023625 0.055987 targetPresent fillerId 0.018832 0.080979 0.152542 rejectId 0.080979 0.163842 0.276836 suspectId 0.158192 0.406780 0.570621 |
- A member variable
dPrimeinDataProcessedis set according to Lineup convention \(d^{\prime}\) is the lowest confidence (most liberal) so
dp.dPrimeis1.767339Showup convention \(d^{\prime}\) is the lowest positive confidence
\(d\) can also be calculated from a signal detection model so
This is calculated from the fit parameters for the fits described in the previous section so
In [X]: mf.d
Out[X]: 1.7976601843420954
Writing results to file
The internal dataframes can be written to either csv or xlsx file format for further analysis. There are four functions belonging to DataProcessed.
writePivotExcelwrites the pivot table to excel
writePivotCsvwrites the pivot table to csv
writeRatesExcelwrites the cummulative rates table to excel
writeRatesCsvwrites the cummulative rates table to csv
The string argument for the functions is the file name.
1 2 3 4 5 6 7 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dp = dr.process() dp.writePivotExcel("test1_pivot.xlsx") dp.writePivotCsv("test1_pivot.csv") dp.writeRatesExcel("test1_rates.xlsx") dp.writeRatesCsv("test1_rates.csv") |
1 2 3 4 5 6 7 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dp <- dr$process() dp$writePivotExcel("./test1_pivot.xlsx") dp$writePivotCsv("./test1_pivot.csv") dp$writeRatesExcel("./test1_rates.xlsx") dp$writeRatesCsv("./test1_rates.csv") |
Designated innocent suspect in target absent (TA) lineups
In TA lineups the suspect ID rate is estimated as the fillerID/lineupSize. In some
experiments a designated innocent suspect is used. This is possible with pyWitness.
The raw data in the
responseTypecolumn will have an extra possible type calleddesignateIdThen all of the analyses will proceed normally, but now the targetAbsent suspectId row will be populated with data from the designated innocent suspect
Sometimes it is useful to check if raw data contains
designateId, this is done by callingdataRaw.isDesignateIdTo convert all
designateIdtofillerIdsone must calldataRaw.removeDesignates()