Advanced tutorials
Bootstrapping uncertainties/confidence limits
To create confidence limits on binned cumulative, statistical measures and parameters, pyWitness uses the bootstrap method. This method takes \(N\) random participants from the original data with replacement. pyWitness can then proceed to compute any quantity (ROC, CAC, pAUC, fit parameters). This is repeated \(M\) times and the distribution of the computed quantity used to calculate a confidence interval with a user definable range.
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
dp = dr.process()
dp.calculateConfidenceBootstrap(nBootstraps=200, cl=95)
dp.printRates()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none())
dp <- dr$process()
dp$calculateConfidenceBootstrap(nBootstraps=as.integer(200), cl=95)
dp$printRates()
After calling calculateConfidenceBootstrap the rates table is populated with the 95% confidence limit
data
confidence
confidence 1 2 3
variable type
cac central 0.861702 0.955614 0.969432
high 0.898148 0.969998 0.982672
low 0.819782 0.934552 0.946858
confidence central 45.873016 74.866469 95.630252
high 47.962794 75.440947 96.258406
low 44.128214 74.301501 94.949277
dprime central 1.975221 1.940776 1.585873
high 2.120760 2.101158 1.834337
low 1.815447 1.753813 1.328038
rf 0.315436 0.428412 0.256152
targetAbsent fillerId 0.284424 0.108352 0.031603
fillerId_high 0.321175 0.134259 0.048269
fillerId_low 0.240872 0.082314 0.016915
rejectId 0.715576 0.521445 0.241535
rejectId_high 0.752949 0.560668 0.277414
rejectId_low 0.672739 0.472804 0.202311
suspectId 0.047404 0.018059 0.005267
suspectId_high 0.053529 0.022377 0.008045
suspectId_low 0.040145 0.013719 0.002819
targetPresent fillerId 0.093960 0.046980 0.013423
fillerId_high 0.118805 0.067776 0.024778
fillerId_low 0.067489 0.025640 0.002278
rejectId 0.286353 0.176734 0.082774
rejectId_high 0.329399 0.211019 0.106384
rejectId_low 0.238167 0.140321 0.055782
suspectId 0.619687 0.438479 0.165548
suspectId_high 0.668216 0.493503 0.196507
suspectId_low 0.567298 0.392393 0.128108
zL central -1.670562 -2.095603 -2.557781
high -1.611559 -2.006968 -2.406877
low -1.749027 -2.205230 -2.768544
zT central 0.304658 -0.154827 -0.971908
high 0.434992 -0.016287 -0.854162
low 0.169499 -0.273088 -1.135387
If a plot function (plotROC, plotCAC) is callled after calling calculateConfidenceBootstrap then
the confidence interval is drawn as error bars, as shown in the ROC plot and CAC plot, respectively, below.
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
dp = dr.process()
dp.calculateConfidenceBootstrap(nBootstraps=200, cl=95)
dp.plotROC()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none())
dp <- dr$process()
dp$calculateConfidenceBootstrap(nBootstraps=as.integer(200), cl=95)
dp$plotROC()
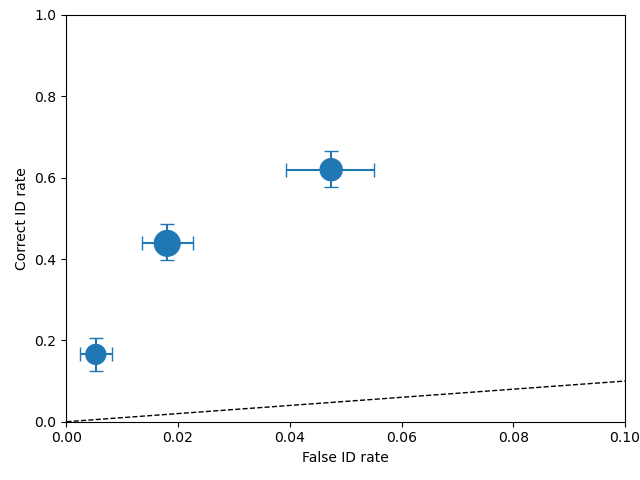
import pyWitness
dr = pyWitness.DataRaw("test1.csv")
dp = dr.process()
dp.calculateConfidenceBootstrap(nBootstraps=200, cl=95)
dp.plotCAC()
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test1.csv")
dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none())
dp <- dr$process()
dp$calculateConfidenceBootstrap(nBootstraps=as.integer(200), cl=95)
dp$plotCAC()
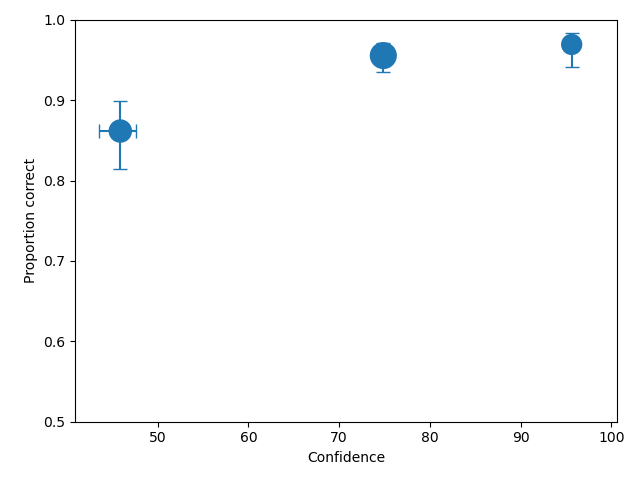
Loading raw data excel format
If the file is in excel format you will need to specify which sheet the raw data is stored in
import pyWitness
dr = pyWitness.DataRaw("test2.xlsx",excelSheet = "raw data")
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test2.xlsx",excelSheet = "raw data")
Transforming data into common format
The raw experimental data does not have to be in the internal format used by pyWitness. As the data is loaded is it possible to replace the name of the data columns and the values stored.
import pyWitness
dr = pyWitness.DataRaw("test2.csv",
dataMapping = {"lineupSize":"lineup_size",
"targetLineup":"culprit_present",
"targetPresent":"present",
"targetAbsent":"absent",
"responseType":"id_type",
"suspectId":"suspect",
"fillerId":"filler",
"rejectId":"reject",
"confidence":"conf_level"}))
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test2.csv",
dataMapping = list("lineupSize"="lineup_size",
"targetLineup"="culprit_present",
"targetPresent"="present",
"targetAbsent"="absent",
"responseType"="id_type",
"suspectId"="suspect",
"fillerId"="filler",
"rejectId"="reject",
"confidence"="conf_level"))
Processing data for two conditions
A single data file might have different experimental condtions. Imagine your data file
has a column labelled Condition and the values for each participant is either Control or
Verbal. To proccess only the Control participants the following options are required
for DataRaw.process()
1 2 3 4 | import pyWitness dr = pyWitness.DataRaw("test2.csv") dr.cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr.process(column="group", condition="Control") |
1 2 3 4 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test2.csv") dr$cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr$process(column="group", condition="Control") |
If you have a file with multiple conditions it is straightforward to make multiple
DataProcessed for each condition, as in the following
1 2 3 4 5 | import pyWitness dr = pyWitness.DataRaw("test2.csv") dr.cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr.process(column="group", condition="Control") dpVerbal = dr.process(column="group", condition="Verbal") |
1 2 3 4 5 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test2.csv") dr$cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl <- dr$process(column="group", condition="Control") dpVerbal <- dr$process(column="group", condition="Verbal") |
Statistical (pAUC) comparision between two conditions
One way to compare pAUC values of two conditions is use the following code on the test2 data. You can check out the script we wrote called pAUCexample.py.
import pyWitness
dr = pyWitness.DataRaw("test2.csv")
dr.cutData(column="previouslyViewedVideo",value=1,option="keep")
dpControl = dr.process(column="group", condition="Control")
dpVerbal = dr.process(column="group", condition="Verbal")
pyw <- import("pyWitness")
dr <- pyw$DataRaw("./test2.csv")
dr$cutData(column="previouslyViewedVideo",value=1,option="keep")
dpControl <- dr$process(column="group", condition="Control")
dpVerbal <- dr$process(column="group", condition="Verbal")
To find the lowest false ID rate from both conditions,
1 2 3 4 5 6 | import pyWitness dr = pyWitness.DataRaw("test2.csv") dr.cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr.process(column="group", condition="Control") dpVerbal = dr.process(column="group", condition="Verbal") minRate = min(dpControl.liberalTargetAbsentSuspectId,dpVerbal.liberalTargetAbsentSuspectId) |
1 2 3 4 5 6 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test2.csv") dr$cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl <- dr$process(column="group", condition="Control") dpVerbal <- dr$process(column="group", condition="Verbal") minRate <- min(dpControl$liberalTargetAbsentSuspectId,dpVerbal$liberalTargetAbsentSuspectId) |
You have to process the data again, with this minRate
1 2 3 4 5 6 7 8 9 10 11 | import pyWitness dr = pyWitness.DataRaw("test2.csv") dr.cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr.process(column="group", condition="Control") dpVerbal = dr.process(column="group", condition="Verbal") minRate = min(dpControl.liberalTargetAbsentSuspectId,dpVerbal.liberalTargetAbsentSuspectId) dpControl = dr.process("group","Control",pAUCLiberal=minRate) dpControl.calculateConfidenceBootstrap(nBootstraps=200) dpVerbal = dr.process("group","Verbal",pAUCLiberal=minRate) dpVerbal.calculateConfidenceBootstrap(nBootstraps=200) dpControl.comparePAUC(dpVerbal) |
1 2 3 4 5 6 7 8 9 10 11 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test2.csv") dr$cutData(column="previouslyViewedVideo",value=1,option="keep") dpControl = dr$process(column="group", condition="Control") dpVerbal = dr$process(column="group", condition="Verbal") minRate = min(dpControl$liberalTargetAbsentSuspectId,dpVerbal$liberalTargetAbsentSuspectId) dpControl = dr$process("group","Control",pAUCLiberal=minRate) dpControl$calculateConfidenceBootstrap(nBootstraps=as.integer(200)) dpVerbal = dr$process("group","Verbal",pAUCLiberal=minRate) dpVerbal$calculateConfidenceBootstrap(nBootstraps=as.integer(200)) dpControl$comparePAUC(dpVerbal) |
To plot the ROC curves, use DataProcess.plotROC
dpControl.plotROC(label = "Control data", relativeFrequencyScale=400)
dpVerbal.plotROC(label = "Verbal data", relativeFrequencyScale=400)
dpControl$plotROC(label = "Control data", relativeFrequencyScale=400)
dpVerbal$plotROC(label = "Verbal data", relativeFrequencyScale=400)
Note
The symbol size is the relative frequency and can be changed by setting dp.plotROC(relativeFrequencyScale = 400)
And your plot will look like this one:
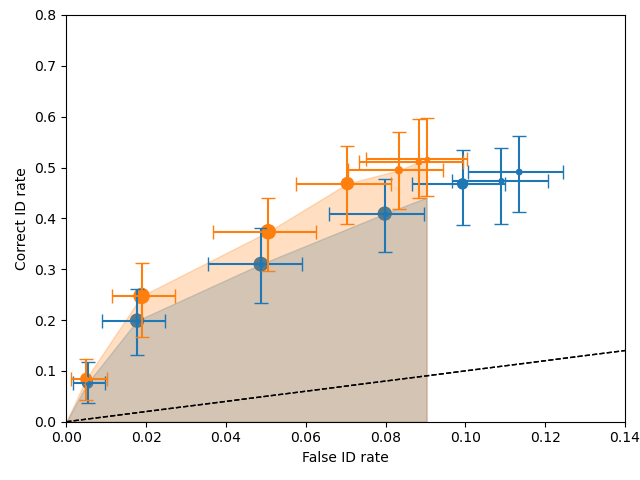
The shaded regions are the pAUCs that were compared. You can see that they both used the same minimum false ID rate. The error bars are 95% confidence intervals. The dashed black line represents chance performance.
Note
The uncertainities can be changed by setting them to .68, for example dpControl.calculateConfidenceBootstrap(nBootstraps=200,cl=68) and dpVerbal.calculateConfidenceBootstrap(nBootstraps=200,cl=68)
Loading processed data
You might already have processed the raw data, or you only have a table of data. It is possible to load a file to perform model fits etc. The processed data need to be in the following CSV format. This is basically the same format as the pivot table stored in DataProcessed.
confidence |
0 |
10 |
20 |
30 |
40 |
50 |
60 |
70 |
80 |
90 |
100 |
targetAbsent fillerId |
2 |
7 |
5 |
8 |
10 |
20 |
26 |
20 |
14 |
8 |
6 |
targetAbsent rejectId |
2 |
5 |
5 |
6 |
9 |
24 |
35 |
56 |
68 |
43 |
64 |
targetPresent fillerId |
0 |
0 |
2 |
3 |
5 |
6 |
5 |
10 |
5 |
4 |
2 |
targetPresent rejectId |
3 |
1 |
0 |
6 |
10 |
20 |
9 |
19 |
23 |
16 |
21 |
targetPresent suspectId |
2 |
1 |
4 |
4 |
10 |
18 |
43 |
68 |
54 |
33 |
41 |
Note
If the targetAbsent suspectId row is not present it is estimated by (targetAbsent fillerId)/lineupSize
The data are stored in data/tutorials/test1_processed.csv
1 2 | import pyWitness dp = pyWitness.DataProcessed("test1_processed.csv", lineupSize = 6) |
1 2 | pyw <- import("pyWitness") dp = pyw$DataProcessed("./test1_processed.csv", lineupSize = 6) |
Using instances of raw data, processed data and model fits
Using an object orientated approach allows multiple instances (objects) to be created and manipulated. This allows many different data file variations on the processed data and model fits to be manipulated simultanuously in a single Python session.
A good example is collapsing data, one might want to check the effect of rebinning the data. In the following example,
the test1.csv is processed twice, once with the original binning (dr1 and dp1) and one with 3 confidence bins
(dr2 and dp2)
import pyWitness
dr1 = pyWitness.DataRaw("test1.csv")
dr2 = pyWitness.DataRaw("test1.csv")
dr2.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels=None)
dp1 = dr1.process()
dp2 = dr2.process()
dp1.plotCAC()
dp2.plotCAC()
pyw <- import("pyWitness")
dr1 <- pyw$DataRaw("./test1.csv")
dr2 <- pyw$DataRaw("./test1.csv")
dr2$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none())
dp1 <- dr1$process()
dp2 <- dr2$process()
dp1$plotCAC()
dp2$plotCAC()
Overlaying plots
In general, each plotXXX function does not create a canvas, so to overlay plots the functions need to be called
sequentially in order.
To make a legend the plots need to be given a label. So this example is the same as the
1 2 3 4 5 6 7 8 9 10 11 12 13 14 | import pyWitness dr1 = pyWitness.DataRaw("test1.csv") dr2 = pyWitness.DataRaw("test1.csv") dr2.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels=None) dp1 = dr1.process() dp2 = dr2.process() dp1.plotCAC(label = "11 bins") dp2.plotCAC(label = "3 bins") import matplotlib.pyplot as _plt _plt.legend() |
pyw <- import("pyWitness")
dr1 <- pyw$DataRaw("./test1.csv")
dr2 <- pyw$DataRaw("./test1.csv")
dr2$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none())
dp1 <- dr1$process()
dp2 <- dr2$process()
dp1$plotCAC(label="11 bins")
dp2$plotCAC(label = "3 bins")
mpl$pyplot$legend()
invisible(mpl$pyplot$ylim(0.50,1.00))
After overlaying plots it maybe important to change the plot axis ranges this can be done with xlim and ylim
xlim(0,100)
ylim(0.50,1.00)
invisible(mpl$pyplot$xlim(0,100))
invisible(mpl$pyplot$ylim(0.50,1.0))
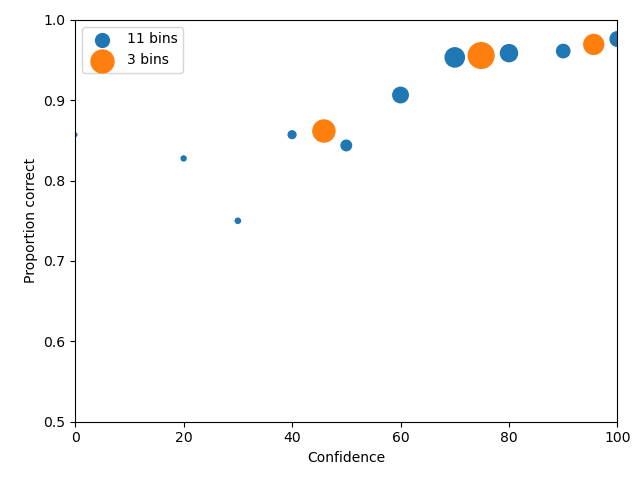
Generating data from signal detection model
Raw and processed data can be generated simply from a signal detection model.
1 2 3 4 5 6 7 8 | import pyWitness dr = pyWitness.DataRaw("test1.csv") dr.collapseContinuousData(column = "confidence",bins = [-1,60,80,100],labels=None) dp = dr.process() mf = pyWitness.ModelFitIndependentObservation(dp, debug=True) mf.setEqualVariance() mf.fit() dr1 = mf.generateRawData(nGenParticipants=10000) |
1 2 3 4 5 6 7 8 | pyw <- import("pyWitness") dr <- pyw$DataRaw("./test1.csv") dr$collapseContinuousData(column = "confidence",bins = c(-1,60,80,100),labels=py_none()) dp <- dr1$process() mf <- pyw$ModelFitIndependentObservation(dp, debug=TRUE) mf$setEqualVariance() mf$fit() dr1 = mf$generateRawData(nGenParticipants=10000) |
dr1 is a DataRaw object and is simulated data for 10,000 participants. dr1 can be used for any
pyWitness analysis so ROC, CAC, pAUC, etc. The raw data can also be written to disk to either preserve and/or
share with colleagues.
1 2 | dr1.writeCsv("fileName.csv") dr1.writeExcel("fileName.xlsx") |
1 2 | dr1$writeCsv("./fileName.csv") dr1$writeExcel("./fileName.xlsx") |
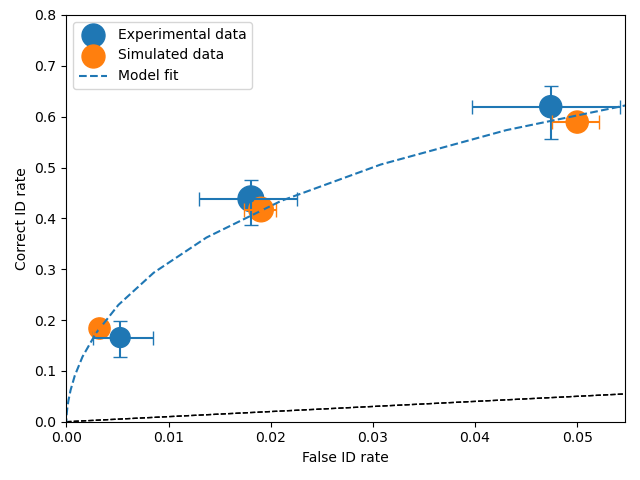
Having performed a fit on dr and generated dr1 a synthetic dataset
# Need to process the synthetic data
dp1 = dr1.process()
# calculate uncertainties using bootstrap
dp.calculateConfidenceBootstrap()
dp1.calculateConfidenceBootstrap()
# plot ROCs
dp.plotROC(label="Experimental data")
dp1.plotROC(label="Simulated data")
mf.plotROC(label="Model fit")
import matplotlib.pyplot as _plt
_plt.legend()
# Need to process the synthetic data
dp1 <- dr1$process()
# calculate uncertainties using bootstrap
dp$calculateConfidenceBootstrap()
dp1$calculateConfidenceBootstrap()
# plot ROCs
dp$plotROC(label="Experimental data")
dp1$plotROC(label="Simulated data")
mf$plotROC(label="Model fit")
mpl$pyplot$legend()
mpl$pyplot$show()
Power analysis
By having the ability to generate data from a model it is possible to vary the number of generated participants. This is not too dissimilar to bootstrapping. Instead of generating new samples (with replacement) from the data, new samples with variable numbers of participants is possible. For each sample all the analysis can be performed and dependence on sample size can be explored.
for nGen in numpy.linspace(500, 5000, 9+1) :
drSimulated = mf.generateRawData(nGenParticipants = nGen)
dpSimulated = drSimulated.process()
dpSimulated.calculateConfidenceBootstrap(nBootstraps=2000)
print(nGen, dpSimulated.liberalTargetAbsentSuspectId,dpSimulated.pAUC, dpSimulated.pAUC_low, dpSimulated.pAUC_high)
for (nGen in list(500,1000,1500)) {
drSimulated <- mf$generateRawData(nGenParticipants = nGen)
dpSimulated <- drSimulated$process()
dpSimulated$calculateConfidenceBootstrap(nBootstraps=as.integer(2000))
print(nGen, dpSimulated$liberalTargetAbsentSuspectId,dpSimulated$pAUC, dpSimulated$pAUC_low, dpSimulated$pAUC_high)
}